The 20 Amino Acids and Their Role in Protein Structures
Overview
Each of the 20 most common amino acids has specific chemical characteristics and a unique role in protein structure and function. Based on the propensity of the side chains to be in contact with water (polar environment), amino acids can be classified into three groups:
1. Those with polar side chains.
2. Those with hydrophobic side chains.
3. Those with charged side chains.
Below we look at each of these classes and briefly discuss their role in protein structure and function.
Each of the 20 most common amino acids has specific chemical characteristics and a unique role in protein structure and function. Based on the propensity of the side chains to be in contact with water (polar environment), amino acids can be classified into three groups:
1. Those with polar side chains.
2. Those with hydrophobic side chains.
3. Those with charged side chains.
Below we look at each of these classes and briefly discuss their role in protein structure and function.
Polar amino acids
When considering polarity, some amino acids are straightforward to define as polar, while in other cases, we may encounter disagreements. For example, serine (Ser, S), threonine (Thr, T), and tyrosine (Tyr, Y) are polar since they carry a hydroxylic (-OH) group. Furthermore, this group can form a hydrogen bond with another polar group by donating or accepting a proton (a table showing donors and acceptors in polar and charged amino acid side chains can be found at the FoldIt site.
Tyrosine is also involved in metal binding in many enzymatic sites. Asparagine (Asn, N) and glutamine (Gln, Q) also belong to this group and may donate or accept a hydrogen bond. Histidine (His, H), on the other hand, depending on the environment and pH, can be polar or carry a charge. It has two –NH groups with a pKa value of around 6. When both groups are protonated, the side chain has a charge of +1. Within protein molecules, the pKa may be modulated by the environment so that the side chain may give away a proton and become neutral or accept a proton, becoming charged. This ability makes histidine useful in enzyme active sites when the chemical reaction requires proton extraction. The aromatic amino acids tryptophan (Trp, W) and Tyr and the non-aromatic methionine (Met, M) are sometimes called amphipathic due to their ability to have both polar and nonpolar character. In protein molecules, these residues are often found close to the interface between a protein and solvent. We should also note here that the side chains of histidine, and tyrosine, together with the hydrophobic phenylalanine and tryptophan, can also form weak hydrogen bonds of the types OH−π and CH−O, using electron clouds within their ring structures. For a discussion of OH−π and CH−O kinds of hydrogen bonds, see Scheiner et al., 2002. A characteristic feature of aromatic residues is that they are often found within the core of a protein structure, with their side chains packed against each other. They are also highly conserved within protein families, with Trp having the highest conservation rate.
Tyrosine is also involved in metal binding in many enzymatic sites. Asparagine (Asn, N) and glutamine (Gln, Q) also belong to this group and may donate or accept a hydrogen bond. Histidine (His, H), on the other hand, depending on the environment and pH, can be polar or carry a charge. It has two –NH groups with a pKa value of around 6. When both groups are protonated, the side chain has a charge of +1. Within protein molecules, the pKa may be modulated by the environment so that the side chain may give away a proton and become neutral or accept a proton, becoming charged. This ability makes histidine useful in enzyme active sites when the chemical reaction requires proton extraction. The aromatic amino acids tryptophan (Trp, W) and Tyr and the non-aromatic methionine (Met, M) are sometimes called amphipathic due to their ability to have both polar and nonpolar character. In protein molecules, these residues are often found close to the interface between a protein and solvent. We should also note here that the side chains of histidine, and tyrosine, together with the hydrophobic phenylalanine and tryptophan, can also form weak hydrogen bonds of the types OH−π and CH−O, using electron clouds within their ring structures. For a discussion of OH−π and CH−O kinds of hydrogen bonds, see Scheiner et al., 2002. A characteristic feature of aromatic residues is that they are often found within the core of a protein structure, with their side chains packed against each other. They are also highly conserved within protein families, with Trp having the highest conservation rate.
Hydrophobic amino acids
The hydrophobic amino acids include alanine (Ala, A), valine (Val, V), leucine (Leu, L), isoleucine (Ile, I), proline (Pro, P), phenylalanine (Phe, F) and cysteine (Cys, C). These residues typically form the hydrophobic core of proteins, which is isolated from the polar solvent. The side chains within the core are tightly packed and participate in van der Waals interactions, which are essential for stabilizing the structure. In addition, Cys residues are involved in three-dimensional structure stabilization through the formation of disulfide (S-S) bridges, which sometimes connect different secondary structure elements or different subunits in a complex. Another essential function of Cys is metal binding, sometimes in enzyme active sites and sometimes in structure-stabilizing metal centers. Also, in the case of Cys, some disagreement exists on its assignment to the hydrophobic group. For example, according to some schemes, it is found in the interior due to its high reactivity (Fiser et al., 1996), while others consider it polar (REF). The image below showing the distribution of buried/exposed fractions of amino acids in protein molecules suggests that Cys is rather exposed.
Charged amino acids
The charged amino acids at neutral pH (around 7.4) carry a single charge in the side chain. There are four of them; the two basic ones include lysine (Lys, K) and arginine (Arg, R), with a positive charge at neutral pH. The two acidic residues include aspartate (Asp, D) and glutamate (Glu, E), which carry a negative charge at neutral pH. A so-called salt bridge is often formed by the interaction of closely located positively and negatively charged side chains. Such bridges are often involved in stabilizing three-dimensional protein structure, especially in proteins from thermophilic organisms (organisms that live at elevated temperatures, up to 80-90 C, or even higher, REF). The binding of positively charged metal ions is another function of the negatively charged carboxylic groups of Asp and Glu. Metalloproteins and the role of metal centers in protein function is a fascinating field of structural biology research.
Glycine & proline
Glycine (Gly), one of the common amino acids, does not have a side chain and is often found at the surface of proteins within loop or coil regions (regions without defined secondary structure), providing high flexibility to the polypeptide chain. This flexibility is required in sharp polypeptide turns in loop structures. Proline, although considered hydrophobic, is also found at the surface, presumably due to its presence in turn and loop regions. In contrast to Gly, which provides the polypeptide chain high flexibility, Pro provides rigidity by imposing certain torsion angles on the segment of the structure (Morgan & Rubenstein, 2013). The reason for this is that its side chain makes a covalent bond with the main chain, which constrains the phi-angle of the polypeptide in this location (see the section of the Ramachandran plot). Sometimes Pro is called a helix breaker since it is often found at the end of α-helices. The importance of Gly and Pro in protein folding has been discussed by (Krieger et al., 2005).
The three-letter and one-letter codes for common amino acids
Charged amino acids (side chains often form salt bridges, in RED mnemonic rules to remember the one-letter code):
• Arginine - Arg - R
• Lysine - Lys - K
• Aspartic acid - Asp - D (AsparDic acid - hard t sound like D)
• Glutamic acid - Glu - E (GluEtamic acid - feels like it is almost an E there)
Polar amino acids (form hydrogen bonds as proton donors or acceptors):
• Glutamine - Gln - Q (Qlutamine - soft G)
• Asparagine - Asn - N (AsparagiNe - clear N)
• Histidine - His - H
• Serine - Ser - S
• Threonine - Thr - T
• Tyrosine - Tyr - Y
• Cysteine - Cys - C
Amphipathic amino acids (often found at the surface of proteins or lipid membranes, sometimes also classified as polar):
• Tryptophan - Trp - W (the largest side chain and the largest letter)
• Tyrosine - Tyr - Y
• Methionine - Met - M (may function as a ligand to metal ions)
Hydrophobic amino acids (normally buried inside the protein core):
• Alanine - Ala - A
• Isoleucine - Ile - I
• Leucine - Leu - L
• Methionine - Met - M
• Phenylalanine - Phe - F
• Valine - Val - V
• Proline - Pro - P
• Glycine - Gly - G
The distribution of amino acids in proteins
The preferred location of different amino acids in protein molecules can be quantitatively characterized by calculating the extent to which an amino acid is buried in the structure or exposed to solvent. The image below provides an overview of the distribution of the different amino acids within protein molecules. While hydrophobic amino acids are mostly buried, a smaller number of polar side chains are buried, while charged residues are mostly exposed.
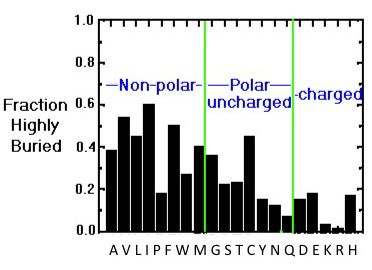
Buried/exposed fraction of amino acids within protein molecules.
The vertical axis shows the fraction of highly buried residues, while the horizontal axis shows the amino acid names in one-letter code. Image from the tutorial by J.E. Wampler,
A discussion on the relationship between surface accessibility and the conservation of amino acids can be found in Fiser et al, 1996.
The vertical axis shows the fraction of highly buried residues, while the horizontal axis shows the amino acid names in one-letter code. Image from the tutorial by J.E. Wampler,
A discussion on the relationship between surface accessibility and the conservation of amino acids can be found in Fiser et al, 1996.
Water & hydrogen bonds
A few words about hydrogen bonds: for a hydrogen bond to be formed, two electronegative atoms (for example, in the case of an α-helix, the amide nitrogen, and the carbonyl oxygen) must interact with the same hydrogen. The hydrogen is covalently attached to one of the atoms (called the hydrogen bond donor) and interacts with the other (the hydrogen bond acceptor). In proteins, all groups capable of forming H-bonds (both main chain and side chain) are usually H-bonded to each other or to water molecules (REF). Due to their electronic structure, water molecules may accept two hydrogen bonds and donate 2, thus being simultaneously engaged in a total of 4 hydrogen bonds. Water molecules may also stabilize protein structures by making hydrogen bonds with the main chain or side chain groups in proteins and even mediating the interactions of different protein groups by linking them to each other by hydrogen bonds. In addition, water is often involved in ligand binding to proteins, mediating ligand interactions with polar or charged side chains or main chain atoms. It is helpful to remember that the energy of a hydrogen bond depends on the distance between the donor and the acceptor and the angle between them and is in the range of 2-10 kcal/mol. The work by Hubbard & Haider, 2010 can be recommended for more detailed information on hydrogen bonds.




