Structural Biology & Bioinformatics: Tools, Databases & Applications
This website offers a guide to structural biology and bioinformatics, highlighting their application in understanding protein structure and function. It includes an overview of sequence and structure bioinformatic databases, tools for amino acid sequence analysis, sequence-structure relationships, and experimental methods in structural biology. Additionally, it introduces protein crystallization and X-ray crystallography, along with basic principles of structure-based drug design.

Amino Acid Sequence Databases
Overview: Sequence analysis
The twenty most common amino acids.
Databases.
Substitution Matrices.
Sequence Alignment.
Tutorial.
Protein Structure & Structure Databases
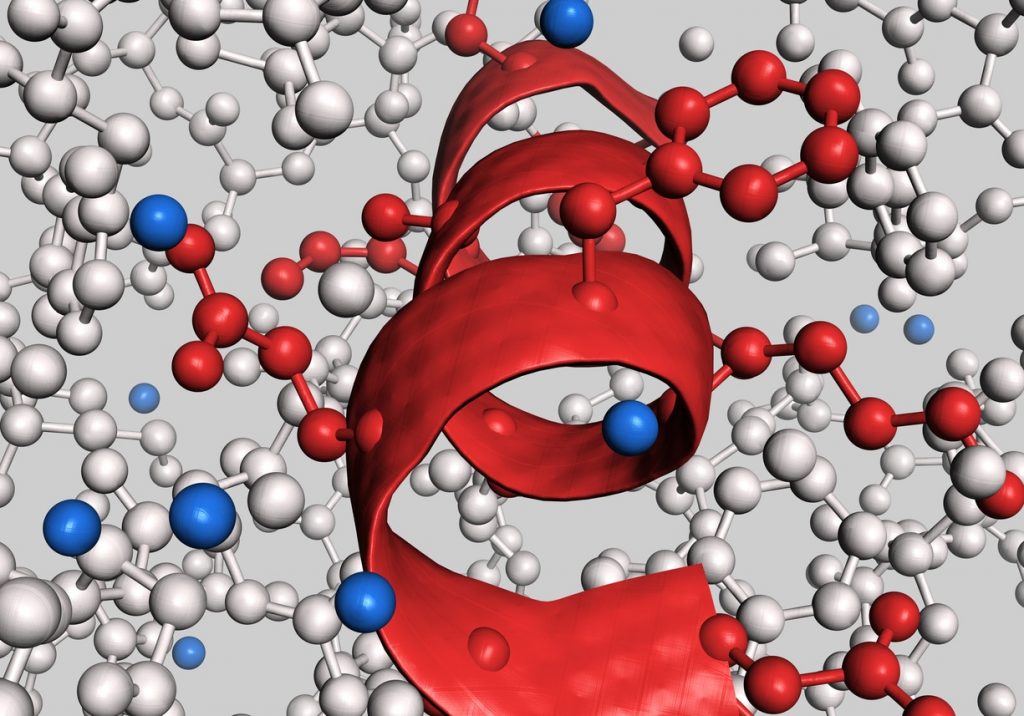
Overview: The four levels of protein structure.
Torsion angles & Ramachandran plot.
Secondary structure.
Domains, classification, databases.

Experimental Methods
Overview: Experimental methods
Crystallization: sitting and hanging drops.
Protein crystallography.
Protein structure quality assessment
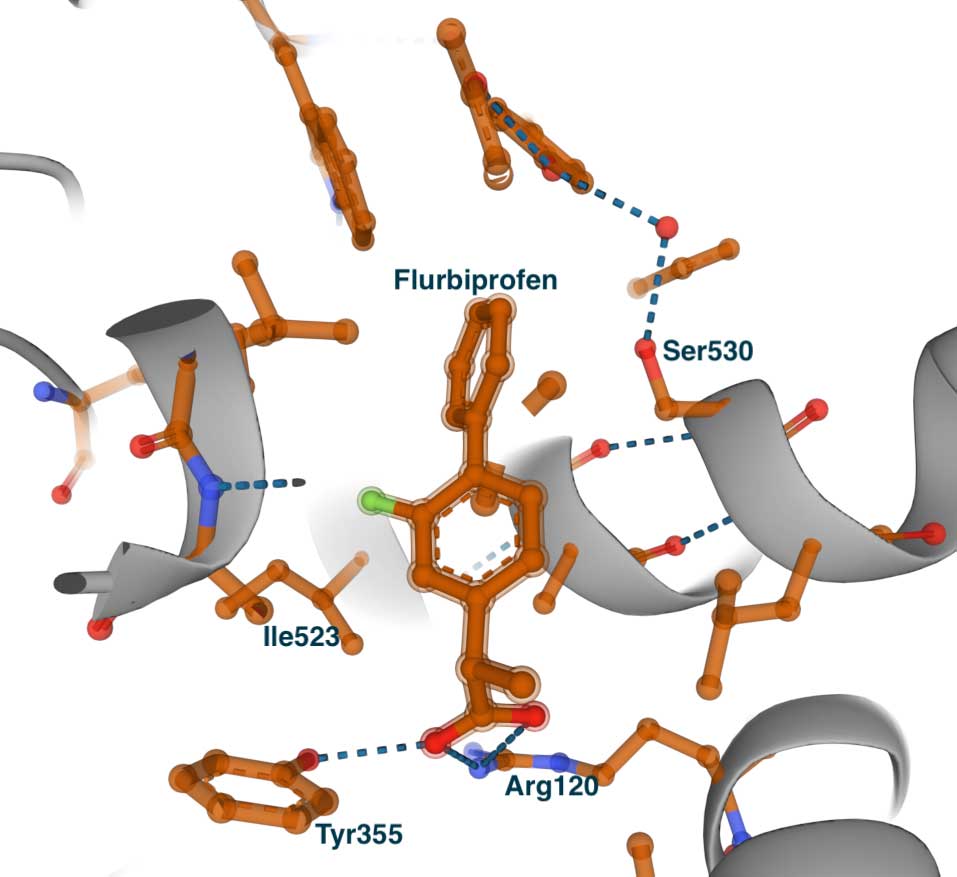
Structure-Based Drug Design
Introduction
Hit identification
Fragment screening
Fragment-based drug design.
The content primarily targets biochemistry and biomedical students interested in structural biology. It is based on a structural biology course that I used to run at Lund University. Just to let you know, this website does not offer a complete course. The complete system consists of lectures, tutorials, and practical exercises, all essential for understanding the principles of structural biology and protein structure and function.
Visit my blog at SARomics.com:



Note for visitors
Please note that the site is updated several times a year, which means it is not updated continuously! I’m very sorry for any inconvenience. The updates may address minor design issues, links present on the site, and more. Additionally, I regret to inform you that I cannot assist with your projects due to time constraints. If you have questions about the website or any comments or suggestions, please feel free to contact me using the contact form.

The content is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.

