Introduction to Protein Structure: Structural Levels, Domains, Motifs and Folds
Overview
To understand the basic principles of three-dimensional protein structure and the potential of their use in various applications in academia and industry, we first need to look at the big picture by defining the four levels of protein structure. The structural levels are mutually dependent on each. They create a complex network of interactions between hundreds and thousands of protein atoms, often involving solvent molecules, various ligands, and metal atoms.
To understand the basic principles of three-dimensional protein structure and the potential of their use in various applications in academia and industry, we first need to look at the big picture by defining the four levels of protein structure. The structural levels are mutually dependent on each. They create a complex network of interactions between hundreds and thousands of protein atoms, often involving solvent molecules, various ligands, and metal atoms.
Primary, secondary, and tertiary structure
The first basic level is the amino acid sequence. The 20 most common amino acids found in proteins are joined together into a polypeptide chain during the process of protein synthesis, catalyzed by the ribosome. To a large extent, the amino acid sequence defines the secondary structure (α-helices and β-sheets) and of course the tertiary structure (domains and folds) of proteins. However, in many cases, we also need to consider the effect of the local environment on structure stabilization. The most obvious example is that of membrane proteins, a considerable part of which is embedded into the hydrophobic environment of membranes. As a result, they lose their native structure outside the membrane and form large aggregates. For this reason, during purification, we need detergent to keep these proteins in a soluble state.
The tertiary structure is defined by the arrangement of the secondary structure elements in space, a fold. The fold is a characteristic feature of the tertiary structure. The currently known three-dimensional protein structures have been classified into around 1300 unique folds. In a later chapter, we will discuss some examples of these folds and the CATH databases where folds and domains are classified, and the relationships between the fold and protein evolutionary origin are revealed.
The tertiary structure is defined by the arrangement of the secondary structure elements in space, a fold. The fold is a characteristic feature of the tertiary structure. The currently known three-dimensional protein structures have been classified into around 1300 unique folds. In a later chapter, we will discuss some examples of these folds and the CATH databases where folds and domains are classified, and the relationships between the fold and protein evolutionary origin are revealed.
Domains, folds & motifs
A domain is defined as an independent folding unit because it may often be cloned, expressed, and purified separately from other domains of a multi-domain protein, and it will still form the same type of structure. It could even show activity (e.g., small molecule ligand binding, interactions with other proteins, etc.) similar to that the domain has within the original protein. A fold is assigned to each protein domain. While some proteins consist of a single domain, others may contain two or more domains with different folds. Domains with similar folds may or may not be related functionally or evolutionary, although a fold can still be used to trace the evolutionary origin of a protein. On the other hand, there are many examples when fold similarity is difficult to detect at the amino acid sequence level.
In addition to domain conservation, there are other types of conserved structural elements in proteins called structural motifs. These smaller structural units may be present within different and not necessarily evolutionary-related domains. Examples of such motifs include helix-turn-helix motifs, β-hairpins, the Greek key motif, and others. However, these motifs are not considered independent folding units in contrast to domains. More detailed discussion and examples can be found on the page on domains, folds, and motifs.
In addition to domain conservation, there are other types of conserved structural elements in proteins called structural motifs. These smaller structural units may be present within different and not necessarily evolutionary-related domains. Examples of such motifs include helix-turn-helix motifs, β-hairpins, the Greek key motif, and others. However, these motifs are not considered independent folding units in contrast to domains. More detailed discussion and examples can be found on the page on domains, folds, and motifs.
Quaternary structure and oligomers
The following structural level is the quaternary structure (also called oligomeric structure). Two or more polypeptide chains (subunits) build these structures. An oligomer may consist of the same type of subunits (homo-oligomer) or be built up by different protein molecules (hetero-oligomer). Oligomeric structures perform a wide range of functions in cells and often work as molecular machines that use ATP as an energy source. An example of the oligomeric structure of the enzyme Mg-chelatase is shown in the image on the left.
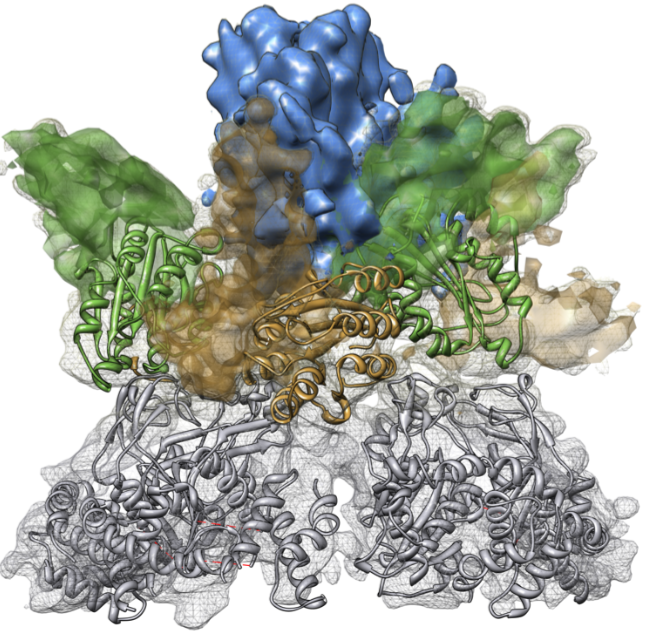
The oligomeric structure of Mg-chelatase
The structure was obtained using single-particle reconstruction from cryo-electron microscopic (cryo-EM) images of the complex. Where appropriate, the available X-ray structure of subunit BchI of the enzyme (shown in ribbon representation) was docked into the EM density. Other domains were modeled based on known structures from other proteins. Published in Lundqvist et al., Structure 2010.
An oligomer is stabilized by interactions between its subunits, such as hydrophobic interactions, hydrogen bonds, and salt bridges. In cases of multi-subunit enzymes, the subunits within the structure often contribute to the formation of the active site or other types of ligand binding sites. An oligomeric complex may also interact with other proteins, forming a so-called transient complex.
Magnesium chelatase, shown in the images above, is involved in chlorophyll biosynthesis. It catalyzes the insertion of magnesium into protoporphyrin IX, which is the first committed step in chlorophyll biosynthesis. About 20 more catalytic reactions are required before the chlorophyll molecule is synthesized.
Mg-chelatase has three different subunits. In bacteriochlorophyll synthesis, they are named BchI, BchD, and BchH. BchI and BchD build a large (about 600 kDa) 2-ring complex shown in the image. The bottom ring is built up by subunit BchI and the top ring by BchD. BchI belongs to the so-called AAA+ family of ATPases. It uses the energy of ATP to drive the Mg-chelatase reaction. It is an example of a molecular machine. In a later section, I will discuss more examples of molecular machines.
Magnesium chelatase, shown in the images above, is involved in chlorophyll biosynthesis. It catalyzes the insertion of magnesium into protoporphyrin IX, which is the first committed step in chlorophyll biosynthesis. About 20 more catalytic reactions are required before the chlorophyll molecule is synthesized.
Mg-chelatase has three different subunits. In bacteriochlorophyll synthesis, they are named BchI, BchD, and BchH. BchI and BchD build a large (about 600 kDa) 2-ring complex shown in the image. The bottom ring is built up by subunit BchI and the top ring by BchD. BchI belongs to the so-called AAA+ family of ATPases. It uses the energy of ATP to drive the Mg-chelatase reaction. It is an example of a molecular machine. In a later section, I will discuss more examples of molecular machines.
Conservation of sequence & structure
Since significant variations in the amino acid sequence within a protein family still yield very similar three-dimensional structures, we say that structure has a higher degree of conservation than sequence. This can be understood if we consider function – for example, binding a ligand, specificity of interactions with other proteins, and structural dynamics – all depend on the three-dimensional structure. Therefore, determining the structure of a protein of unknown function and its subsequent comparison to other known structures in a database may help reveal structural homologs and, ultimately, the protein's function. The principle of structure conservation also allows us to rely on structure prediction and modeling when no experimental structure is available.
An exciting example of structure conservation was provided by the anaerobic cobalt chelatase, an enzyme active in vitamin B12 synthesis (Schubert et al., 1999). Although the protein's function was already known, only the determination of its three-dimensional structure revealed its similarity to the structure of ferrochelatase (Al-Karadaghi et al., 1997), an enzyme active in heme biosynthesis. The reason is that the sequence identity between the two proteins is only 11%, a number much smaller than the so-called "homology threshold" (around 20-25% sequence identity). The similarity between the two structures strongly suggested a common evolutionary origin of the two proteins and a similarity in the mechanism of the enzymatic reaction.
An exciting example of structure conservation was provided by the anaerobic cobalt chelatase, an enzyme active in vitamin B12 synthesis (Schubert et al., 1999). Although the protein's function was already known, only the determination of its three-dimensional structure revealed its similarity to the structure of ferrochelatase (Al-Karadaghi et al., 1997), an enzyme active in heme biosynthesis. The reason is that the sequence identity between the two proteins is only 11%, a number much smaller than the so-called "homology threshold" (around 20-25% sequence identity). The similarity between the two structures strongly suggested a common evolutionary origin of the two proteins and a similarity in the mechanism of the enzymatic reaction.