Torsion Angles and the Ramachandran Plot
Definition
There are two essential torsion (rotation) angles in the polypeptide chain, also called Ramachandran angles (after the Indian physicist who worked on modeling the interactions in polypeptide chains, Ramachandran, GN, et al., J Mol Biol, 7:95-99). These angles describe the rotations of the polypeptide backbone around the bonds between N-Cα (called Phi, φ) and Cα-C (called Psi, ψ) and essentially determine the fold of the protein. An image for the graphics view of the angles is shown below.
There are two essential torsion (rotation) angles in the polypeptide chain, also called Ramachandran angles (after the Indian physicist who worked on modeling the interactions in polypeptide chains, Ramachandran, GN, et al., J Mol Biol, 7:95-99). These angles describe the rotations of the polypeptide backbone around the bonds between N-Cα (called Phi, φ) and Cα-C (called Psi, ψ) and essentially determine the fold of the protein. An image for the graphics view of the angles is shown below.
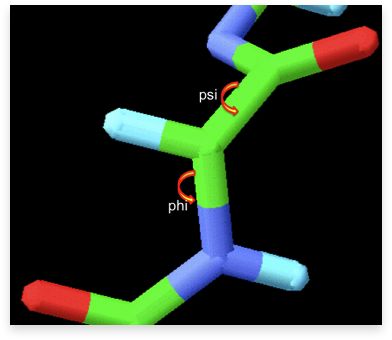
A fragment of a polypeptide chain showing the rotation around the bond that defines the torsion angles φ and ψ as rounded arrows.
The torsion angle (dihedral angle) is defined by three consecutive bonds involving four atoms. The angle describes the rotation of the chain around the middle bond. In proteins, the two torsion angles φ and ψ (Ramachandran angles) describe the rotation around N-Cα and Cα-C bonds, respectively.
The torsion angle (dihedral angle) is defined by three consecutive bonds involving four atoms. The angle describes the rotation of the chain around the middle bond. In proteins, the two torsion angles φ and ψ (Ramachandran angles) describe the rotation around N-Cα and Cα-C bonds, respectively.

Structural biology services by SARomics Biostructures

X-ray Crystallography

NMR spectroscopy

Structure-based drug design
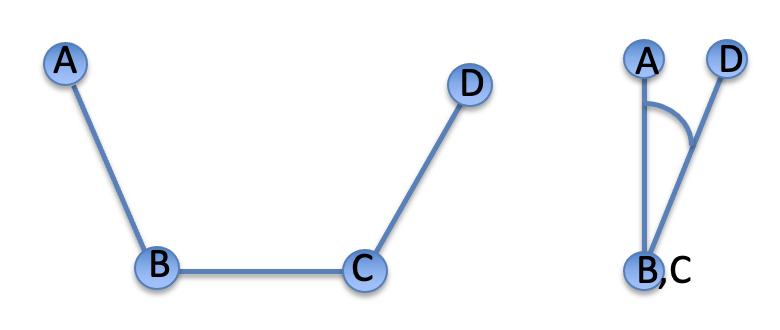
The standard IUPAC definition of a dihedral angle is illustrated in the Figure. A, B, C, and D illustrate the position of the four atoms used to define the dihedral angle. The rotation takes place around the central B-C bond. The view on the right is along the B-C bond with atom A placed at 12 o'clock. The rotation around the B-C bond is described by the A-B-D angle shown on the right. Positive angles correspond to clockwise rotation.
The Ramachandran plot
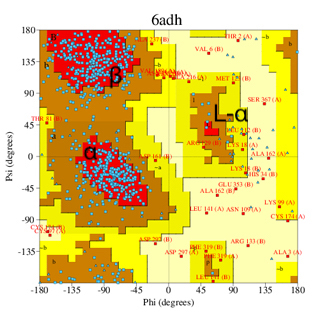
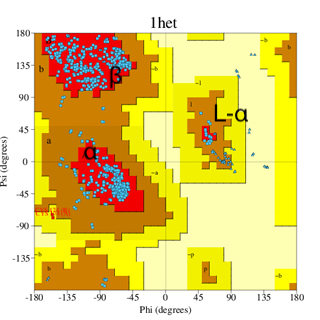
A special way for plotting and visualization of protein torsion angles was introduced by Ramachandran and co-authors and has since then been called the Ramachandran plot. In the Ramachandran plot, we can view the distribution of torsion angles in a protein structure (image below). The horizontal axis on the plot shows φ values, while the vertical shows ψ values. Both horizontal and vertical axes start from -180 and extend to +180. Each dot in the plot corresponds to an amino acid, with its φ and ψ angles. In the images, the regions corresponding to φ and ψ angles of α-helices and β-sheets are marked. The two regions are well separated and occupy different plot areas (marked as α and β). We can also see that the distribution of the torsion angles is different in the two images. The dots are much better clustered in the right image than in the left. What is the reason for this? This depends on the quality of the structures and is discussed below on this page.
Theoretically, the average phi and psi values for α-helices and β-sheets were predicted to cluster around -57, -47, and -80, +150, respectively. However, for actual experimental structures, these values were found to be different. For those who may be interested in details of the distribution of torsion angles in a Ramachandran plot, I can recommend the paper by Hovmöller et al., 2002, which provides an excellent discussion of the subject.
Theoretically, the average phi and psi values for α-helices and β-sheets were predicted to cluster around -57, -47, and -80, +150, respectively. However, for actual experimental structures, these values were found to be different. For those who may be interested in details of the distribution of torsion angles in a Ramachandran plot, I can recommend the paper by Hovmöller et al., 2002, which provides an excellent discussion of the subject.
A Ramachandran plot showing the distribution of the torsion angles of a protein refined at two different resolutions.
The image on the left (PDB code 6adh) corresponds to a structure at around 2.9 Å resolution, while on the right, the structure (1het) is at 1.15 Å resolution (much higher resolution). It can be seen that the dots for the high resolution structure (1het) are nicely clustered in the energetically most favorable red regions of α-helices, β-sheets, and left-handed α-helix L-α. On the other hand, at the lower resolution, many dots are found in energetically less favorable (yellow) regions. This is due to the better quality of the high resolution structure (see text below).
The horizontal axis on the plot shows φ values, while the vertical shows ψ values. Both horizontal and vertical axes start from -180 and extend to +180. The upper red region cluster corresponds to β-structure torsion angles, and the lower red region cluster corresponds to α-helical structures.
Images generated at PDBsum.
The image on the left (PDB code 6adh) corresponds to a structure at around 2.9 Å resolution, while on the right, the structure (1het) is at 1.15 Å resolution (much higher resolution). It can be seen that the dots for the high resolution structure (1het) are nicely clustered in the energetically most favorable red regions of α-helices, β-sheets, and left-handed α-helix L-α. On the other hand, at the lower resolution, many dots are found in energetically less favorable (yellow) regions. This is due to the better quality of the high resolution structure (see text below).
The horizontal axis on the plot shows φ values, while the vertical shows ψ values. Both horizontal and vertical axes start from -180 and extend to +180. The upper red region cluster corresponds to β-structure torsion angles, and the lower red region cluster corresponds to α-helical structures.
Images generated at PDBsum.
The Ramachandran plot and structure quality
The higher resolution of the X-ray data usually results in a higher quality three-dimensional structure. For example, in the Ramachandran plot for the higher resolution structure on the image on the right, it is easy to see that the values of the torsion angles are clustered within the red regions (α- and β-regions). These are called allowed or energetically most favorable regions. Other regions are less favorable and are poorly populated in good-quality structures (yellow color). This results from a steric hindrance – specific rotations around the polypeptide chain will bring atoms too close to each other, creating high energy steric repulsion. For this reason, the Ramachandran plot serves as one of the critical indicators of the quality of a three-dimensional structure – a good quality structure is expected to have the majority, if not all, of its torsion angles within the allowed regions of the plot (image on the right).
However, sometimes we may find amino acids with "wrong" torsion angles, which may be for a good reason. The strain (high energy) created in a structure by such conformations may have functional significance (Pal & Chakrabarti, 2002) and may even be conserved within a protein family!
Another exception from the principle of clustering around the α- and β-regions is provided by glycine. Glycine residues do not have a side chain, which allows a higher flexibility in regions of the polypeptide chain where G is located (usually loops, which explains the relatively high exposure of Gly in protein structures). As a result, the regions may adopt conformations that otherwise are forbidden. As mentioned earlier, proline, in contrast to glycine, fixes the torsion angles at a specific value, very close to that of an extended β-strand. A more detailed discussion of the quality of experimental structures can be found on the page on quality assessment.
However, sometimes we may find amino acids with "wrong" torsion angles, which may be for a good reason. The strain (high energy) created in a structure by such conformations may have functional significance (Pal & Chakrabarti, 2002) and may even be conserved within a protein family!
Another exception from the principle of clustering around the α- and β-regions is provided by glycine. Glycine residues do not have a side chain, which allows a higher flexibility in regions of the polypeptide chain where G is located (usually loops, which explains the relatively high exposure of Gly in protein structures). As a result, the regions may adopt conformations that otherwise are forbidden. As mentioned earlier, proline, in contrast to glycine, fixes the torsion angles at a specific value, very close to that of an extended β-strand. A more detailed discussion of the quality of experimental structures can be found on the page on quality assessment.